Un tipo de secuencia común de Salmonella Enteritidis de origen aviar y de humano con gastroenteritis en Ibagué, Colombia
Resumen
Introducción. Salmonella Enteritidis es una de las mayores causas de salmonelosis en el mundo, siendo los huevos contaminados y la carne de pollo cruda sus principales fuentes de infección. En Ibagué, Colombia, se identificaron los principales serovares circulando en granjas, superficies de huevos y canales de pollo, sin embargo, se desconoce si esos serovares son responsables de gastroenteritis.
Objetivo. Evaluar la relación genética entre aislamientos de Salmonella Enteritidis de aves de corral y humanos con gastroenteritis mediante multilocus sequence typing (MLST).
Materiales y métodos. Se aisló Salmonella spp., de casos clínicos de gastroenteritis (n=110). Se realizó test de sensibilidad antibiótica, seguido de serotipificación y tipificación por medio de MLST y se comparó S. Enteritidis de humanos frente a S. Enteritidis de granjas ponedoras y de huevo comercializado (n=6).
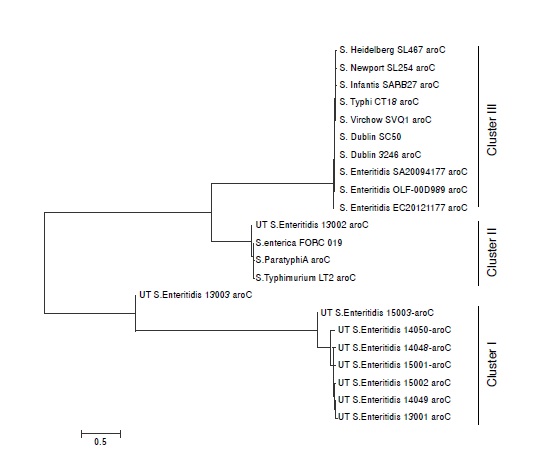
Resultados. Se aislaron 10 cepas de Salmonella spp., a partir de heces de humanos con gastroenteritis. Se obtuvo una prevalencia de Salmonella spp. de 9.09%, siendo S. Enteritidis (n=4), S. Typhymurium (n=2), S. Newport (n=1), S. Grupensis (n=1), S. Uganda (n=1) y S. Braenderup (n=1) los serotipos presentes en pacientes con gastroenteritis. El MLST indico que un tipo de secuencia común (ST11) de S. Enteritidis estuvo presente en todas las tres fuentes y mostraron el mismo patrón de resistencia antibiótica.
Conclusión. S. Enteritidis ST11 constituye un vínculo entre el consumo/manipulación de huevos contaminados y gastroenteritis humana en Ibagué. Son necesarios estudios complementarios para conocer si otros serovares de Salmonella aislados de carne de pollo cruda también se asocian con la gastroenteritis humana.
Descargas
Referencias bibliográficas
Grimont PA, Weill FX. Antigenic formulae of the Salmonella serovars. Ninth edition. Paris, France: Institute Pasteur, WHO; 2007. p. 1-166.
Sanderson KE, Nair S. Taxonomy and species concepts in the genus Salmonella. In: Barrow PA, Methner U, editors. Salmonella in domestic animals. Wallingford: CABI; 2013. p. 1-19. https://doi.org/10.1079/9781845939021.0001
Landgridge GE, Wain J, Nair S. Invasive salmonellosis in humans. EcoSal Plus. 2012;5:1-14. https://doi.org/10.1128/ecosalplus.8.6.2.2
Organización Mundial de Sanidad Animal. Manual de las pruebas de diagnóstico y de las vacunas para los animales terrestres, 2017. Chapter 2.9.8. Salmonellosis. 2010. Accessed: January 22, 2017. Available from: http://www.oie.int/fileadmin/Home/esp/Health_standards/tahm/2.09.08_SALMONELLOSIS.pdf
Centers for Disease Control and Prevention. Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet Surveillance Report for 2012. Atlanta, GA: U. S. Department of Health and Human Services, CDC; 2014. Accessed: January 22, 2017. Available from: https://www.cdc.gov/foodnet/PDFs/2012_annual_report_508c.pdf
European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2016;14:4634-231. https://doi.org/10.2903/j.efsa.2016.4634
Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, et al. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: Results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis. 2011;8:887-900. https://doi.org/10.1089/fpd.2010.0787
Braden CR. Salmonella enterica serotype Enteritidis and eggs: A national epidemic in the United States. Clin Infect Dis. 2006;43:512-7. https://doi.org/10.1086/505973
Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. Outreak-associated Salmonella enterica Serotypes and Food Commodities, United States, 1998-2008. Emerg Infect Dis. 2013;19:1239-44. https://doi.org/10.3201/eid1908.121511
Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al. International Collaboration on Enteric Disease “Burden of Illness” Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882-9. https://doi.org/10.1086/650733
Dekker JP, Frank KM.Salmonella, Shigella, and Yersinia. Clin Lab Med. 2015;35:225-46. https://doi.org/10.1016/j.cll.2015.02.002
Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States - major pathogens. Emerg Infect Dis. 2011;17:7-15. https://doi.org/10.3201/eid1701.P11101
Public Health Agency of Canada. Public Health Notice-Outbreak of Salmonella infections under investigation. March 4, 2016. Accessed: January 22, 2017. Available from: http://www.phac-aspc.gc.ca/phn-sp/2015/salmonella-infantis-eng.php
Public Health England. Research and Analysis, Salmonella infections (fecal specimens) in England and Wales: Laboratory reports 2016. Accessed: January 25, 2017. Available from: https://www.gov.uk/government/publications/salmonella-infections-faecal-specimens-inengland-and-wales-laboratory-reports-2016
European Food Safety Authority. Multi-country outbreak of Salmonella Enteritidis infections associated with consumption of eggs from Germany. EFSA J. 2014;11:646E. https://doi.org/10.2903/sp.efsa.2014.EN-646
European Centre for Disease Prevention and Control. Communicable disease threats report 3-September, 2016. Accessed: January 22, 2017. Available from: www.ecdc.europa.eu
Instituto Nacional de Salud. Boletín Epidemiológico semana 21 de 2015. Dirección de Vigilancia y Análisis del riesgo en Salud Pública. 2015. Accessed: January 25, 2017. Available from: http://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2015%20Boletin%20epidemiologico%20semana%2021.pdf
Rodríguez R, Fandiño C, Donado P, Guzmán L, Verjan N. Characterization of Salmonella from commercial egg-laying hen farms in a central region of Colombia. Avian Dis. 2015;59:59-63. https://doi.org/10.1111/j.1440-1681.2011.05632.x
Mogollón DC, Rodríguez VE, Verjan N. Prevalence and molecular identification of Salmonella isolated from commercialized eggs at Ibagué, Colombia. Rev Salud Anim. 2016;38:164-72.
Rodríguez J, Rondón I, Verjan N. Serotypes of Salmonellain broiler carcasses marketed at Ibagué, Colombia. Rev Bras Ciên Avic. 2015;17:545-52. https://doi.org/10.1590/1516-635X1704545-552
Donado-Godoy P, Gardner I, Byrne A, Leon M, Pérez-Gutiérrez E, Ovalle MV, et al. Prevalence, risk factors, and antimicrobial resistance profiles of Salmonella from commercial broiler farms in two important poultry-producing regions of Colombia. J Food Prot. 2012;75:874-83. https://doi:10.4315/0362-028X.JFP-11-458
Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561-88. https://doi.org/10.1146/annurev.micro.59.030804.121325
Pérez-Lozada M, Cabezas P, Castro-Nallar E, Crandall KA. Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect Genet Evol. 2013;16:38-53. https://doi.org/10.1016/j.meegid.2013.01.009
Thrusfield M. Veterinary epidemiology. Third edition. Ames, IO: Blackwell Publishing; 2007. p. 46-74.
Tindall BJ, Grimont PA, Garrity GM, Euzéby JP. Nomenclature and taxonomy of the genus Salmonella. Int J Syst Evol Microbiol. 200555:521-4. https://doi.org/10.1099/ijs.0.63580-0
Clinical and Laboratory Standards Institute. CLSI publishes new antimicrobial susceptibility testing standards. Wayne: CLSI; 2014.
Sambrook J, Russell D. Molecular cloning: A laboratory manual. Third edition. Cold Spring: Harbor Laboratory Press; 2001. p. 2100.
Rahn K, De Grandis A, Clarke R, McEwen A, Galán J, Ginocchio E, et al. Amplification of an InvA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271-9. https://doi.org/10.1016/0890-8508(92)90002-F
Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8:e1002776. https://doi.org/10.1371/journal.ppat.1002776
Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203-14. https://doi.org/10.1089/10665270050081478
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis, version 6.0. Mol Biol Evol. 2013;30:2725-9. https://doi.org/10.1093/molbev/mst197
Ministerio de la Protección Social. Perfil de riesgo Salmonella spp. (no tifoideas) en pollo entero y en piezas. Bogotá: Unidad de Evaluación de Riesgos para la Inocuidad de los Alimentos, UERIA; 2011.
Barua H, Biswas PK, Talukder KA, Olsen KE, Christensen JP. Poultry as a possible source of non-typhoidal Salmonella enterica serovars in humans in Bangladesh. Vet Microbiol. 2014;168:372-80. https://doi.org/10.1016/j.vetmic.2013.11.020
Pulido M, Sánchez R, Guard J, Do Nascimento V. Presence of Salmonella Enteritidis and Salmonella Gallinarum in commercial laying hens diagnosed with fowl typhoid disease in Colombia. Avian Dis. 2014;58:165-70. https://doi.org/10.1637/10598-062613-Case.1
Favier GI, Lucero-Estrada CS, Lazarte-Otero V, Escudero ME. Prevalence, antimicrobial susceptibility, and molecular characterization by PCR and pulsed field gel electrophoresis (PFGE) of Salmonella spp. isolated from foods of animal origin in San Luis, Argentina. Food Control. 2013;29:49-54. https://doi.org/10.1016/j.foodcont.2012.05.056
Campioni F, Davis M, Medeiros MI, Falcão JP, Shah DH. MLVA typing reveals higher genetic homogeneity among S. Enteritidis strains isolated from food, humans and chickens in Brazil in comparison to the North American Strains. Int J Food Microbiol. 2013;162:174-81. https://doi.org/10.1016/j.ijfoodmicro.2013.01.008
Martelli F, Davies RH. Salmonella serovars isolated from table eggs: An overview. Food Res Int.2012;45:745-54. https://doi.org/10.1016/j.foodres.2011.03.054
Howard ZR, O’Bryanb CA, Crandallb PG, Rickeb SC. Salmonella Enteritidis in shell eggs: Current issues and prospects for control. Food Res Int. 2012;45:755-64. https://doi.org/10.1016/j.foodres.2011.04.030
Rodrigue DC, Tauxe RV, Rowe B. International increase in Salmonella Enteritidis: A new pandemic? Epidemiol Infect. 1990;105:21-7.
Noda T, Murakami K, Asai T, Etoh Y, Ishihara T, Kuroki T, et al. Multi-locus sequence typing of Salmonella enterica subsp. enterica serovar Enteritidis strains in Japan between 1973 and 2004. Acta Vet Scand. 2011;53:38. https://doi.org/10.1186/1751-0147-53-38
Ji-Yeon H, Jung-Whan C, Jun-Ho P, Moo-Sang K, Young-Hee O, In-Soo C, et al. A comparison of subtyping methods for differentiating Salmonella enteric serovar Enteritidis isolates obtained from food and human sources. Osong Public Health Res Perspect. 2013;4:27-33. https://doi.org/10.1016/j.phrp.2012.12.005
Campioni F, Pitondo-Silva A, Bergamini AM, Falcao JP. Comparison of four molecular methods to type Salmonella Enteritidis strains. APMIS. 2015;123:422-6. https://doi.org/10.1111/apm.12367
Milanez GP, Nascimento LC, Tirabassi AH, Zuanaze M, Rodrigues DP, Pereira GAG, et al. Whole-genome sequence of Salmonella enterica serovar Enteritidis phage type 4, isolated from a Brazilian poultry farm. Genome Announc. 2016;4:e00340-16. https://doi.org/10.1128/genomeA.00340-16
Retamal P, Fresno M, Dougnac C, Gutiérrez S, Gornall V, Vidal R, et al. Genetic and phenotypic evidence of the Salmonella enterica serotype Enteritidis human-animal interface in Chile. Front Microbiol. 2015;6:1-10. https://doi.org/10.3389/fmicb.2015.00464
Ghaderi R, Tadayon K, Khaki P, Mosavari N. Iranian clonal population of Salmonella enterica serovar Enteritidis, characterized by multi-locus sequence typing (MLST) method. Iran J Microbiol. 2015;7:251-9.
Murgia M, Bouchrif B, Timinouni M, Al-Qahtani A, Al-Ahdal MN, Cappuccinelli P, et al. Antibiotic resistance determinants and genetic analysis of Salmonella enterica isolated from food in Morocco. Int J Food Microbiol. 2015;215:31-9. https://doi.org/10.1016/j.ijfoodmicro.2015.08.003
Kim Y, Baea IK, Jeong SH, Leeb CH, Lee HK, Ahnd J, et al. Occurrence of IncFII plasmids carrying the blaCTX-M-15 gene in Salmonella enterica serovar Enteritidis sequence type 11 in Korea. Diagn Microbiol Infect Dis. 2011;71:171-3. https://doi.org/10.1016/j.diagmicrobio.2011.05.004
Hwang JH, Shin GW, Lee CS. Community-onset pyomyositis caused by a Salmonella enterica serovar Enteritidis sequence type 11 strain producing CTX-M-15 extendedspectrum - lactamase. J Clin Microbiol. 2015;53:1439-41. https://doi.org/10.1128/JCM.03097-14
Jones-Dias D, Clemente L, Egas C, Froufe H, Sampaio DA, Vieira L, et al. Salmonella enteritidis isolate harboring multiple efflux pumps and pathogenicity factors, shows absence of o antigen polymerase gene. Front Microbiol. 2016;7:1130. https://doi.org/10.3389/fmicb.2016.01130
Barco L, BarrucciF, Elmerdahl JO, Ricci A. Salmonella source attribution based on microbial subtyping. Int J Food Microbiol. 2013;163:193-203.
Algunos artículos similares:
- Oscar G. Gómez, Vacuna atenuada de Salmonella como vector de antígenos heterólogos , Biomédica: Vol. 20 Núm. 2 (2000)
- Johnny Durango, Germán Arrieta, Salim Mattar, Presencia de Salmonella spp. en un área del Caribe colombiano: un riesgo para la salud pública. , Biomédica: Vol. 24 Núm. 1 (2004)
- Zamira E. Soto-Varela, Clara Gilma Gutiérrez, Yurina de Moya, Ramón Mattos, Hernando José Bolívar-Anillo, José Luis Villarreal, Detección molecular de Salmonella spp., Listeria spp. y Brucella spp. en queso artesanal fresco comercializado en Barranquilla: un estudio piloto , Biomédica: Vol. 38 Núm. Sup. 2 (2018): Suplemento 2, Medicina tropical
- Carolina del Carmen Murúa-López, María González-Orozco, Héctor Samuel López-Moreno, Las células dendríticas plasmacitoides evocan la respuesta efectora de los linfocitos T citotóxicos específicos para Salmonella. , Biomédica: Vol. 39 Núm. Supl. 2 (2019): Enfermedades transmisibles en el trópico, agosto
- Nancy Yaneth Flórez , Stefany Alejandra Arévalo , Edna Catering Rodríguez , Jaime Guerrero , Kelly Paola Valverde , Paula Lucía Díaz , Lucy Angeline Montaño, Doris Mabel Gartner , Carolina Duarte , Jaime Enrique Moreno, Brote de Salmonella enterica subsp. enterica serovar Give asociado con enfermedad transmitida por alimentos en Vichada, Colombia, 2015 , Biomédica: Vol. 41 Núm. 1 (2021)
- Edna Catering Rodríguez, Sandra Yamile Saavedra, Lucy Angeline Montaño, Diana Patricia Sossa, Francia Patricia Correa, Jireh Alejandra Vaca, Carolina Duarte, Caracterización de β-lactamasas de espectro extendido en aislamientos clínicos colombianos de Salmonella enterica no tifoidea de 1997 a 2022 , Biomédica: Vol. 43 Núm. 3 (2023)
| Estadísticas de artículo | |
|---|---|
| Vistas de resúmenes | |
| Vistas de PDF | |
| Descargas de PDF | |
| Vistas de HTML | |
| Otras vistas | |